Over the past month, I have been creating graphs using R that visualize influenza lineages by different patient demographic variables. However, after creating some of these graphs and discussing them with Dr. Qin, the head of the lab, we concluded that graphing with more specific sub-lineage data would allow for a better analysis. This means that instead of only distinguishing between two main lineages of influenza, I will be graphing using data that specifies between different influenza sub-clades. Clades and sub-clades are further subdivisions of the main lineages, which tell us even more about the evolution of the virus.
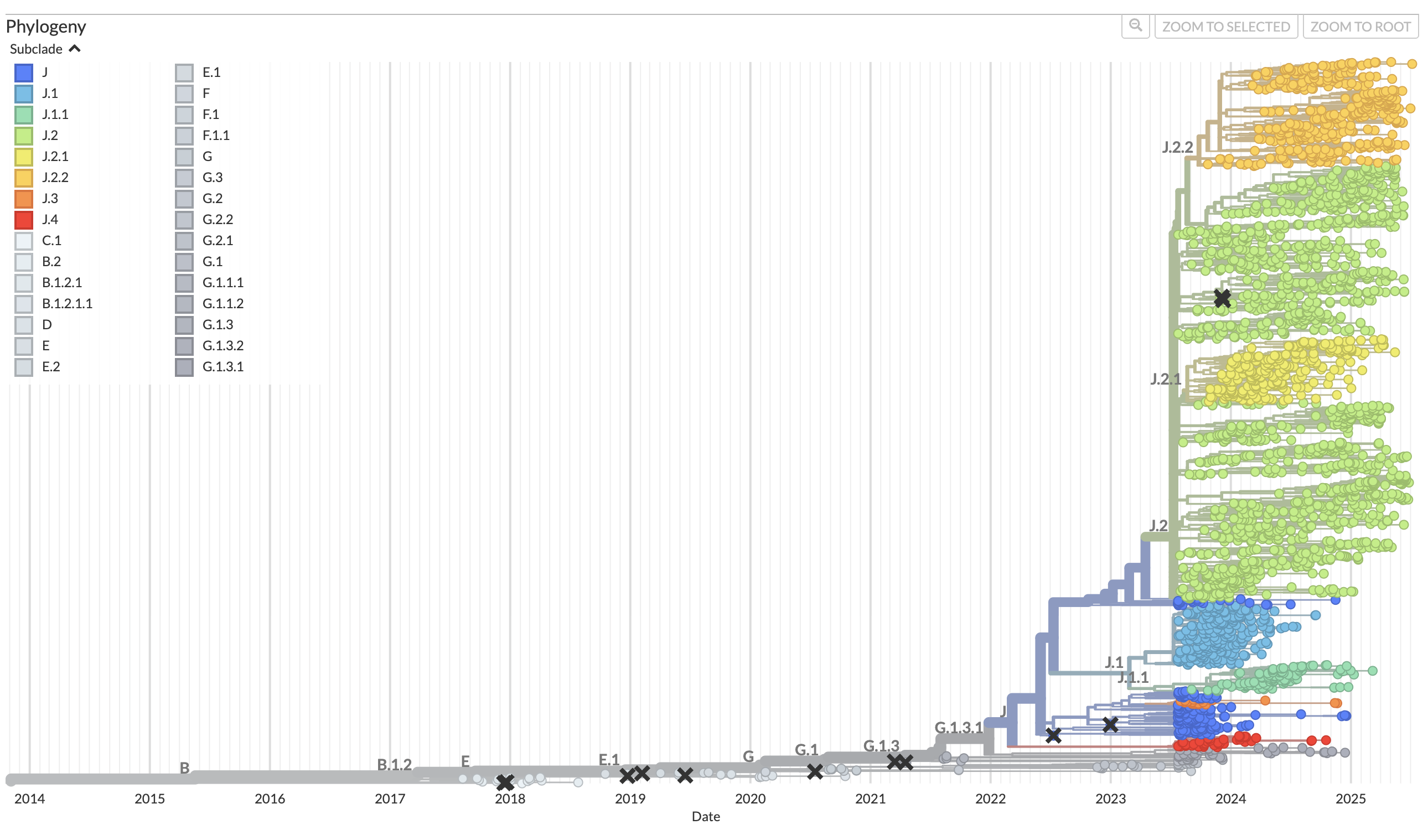
While I continue to work on creating new graphs, Dr. Qin has been introducing me to an important tool called Nextstrain. Nextstrain is a project run by a team who pulls data from a place called GISAID, which is an incredible global database where labs, including our lab, upload their genomic data from respiratory virus samples for rapid and open data sharing. Nextstrain has put together phylogenetic trees of respiratory virus evolution using the open access data from GISAID.
This phylogenetic tree from Nextstrain shows influenza evolution from 2013 to the present day, using 2891 genomes! The different sub-clades are grouped by color. Using this open access data, I will be able to compare our lab’s dataset, which only contains data from the Portland metro area, with data from other locations around the United States, and even around the world! This can help us answer questions about how influenza in Oregon may be evolving differently than influenza in other locations.
Data analysis for this project has felt tedious at times, as there are so many different variables to compare. I have made tangible progress by creating graphs, however there is still much to be done! I am excited to keep working and hopefully come to some conclusions about influenza evolution in the next month.
- EVA VU-STERN '28